ROS-1 rearrangements with at least 12 different partner proteins have been identified in a small subset of lung non-small cell carcinomas (1–2%), which shows susceptibility to tyrosine kinase inhibitors (TKIs) similar to ALK rearranged tumors. ROS-1 is considered a oncogene found on chromosome 6. The exact mechanism of activation of this gene protein product with the various gene rearrangement partners is not understood. The protein function is similar to that of the ALK family, which is why this mutation was studied for possible response to ALK inhibitors (crizotinib).
Recently, ROS-1 mutated tumors have been approved for TKI therapy with identification of a rearrangement by FISH analysis. Like ALK, ROS-1 FISH utilizes a break apart probe to identify the presence of a gene rearrangement. Other successful modalities for identification of ROS-1 rearrangements include ‘next generation’ sequencing (NGS) and immunohistochemistry.
Immunohistochemistry (IHC) has been studied as an alternative to FISH as a screening modality. Based on multiple studies, the sensitivity of IHC appears to be near 100% with the specificity of at least 92%. These studies were performed using the D4D6 rabbit monoclonal antibody clone (Cell Signaling Technology, Danvers, Massachusetts).
Stain Interpretation
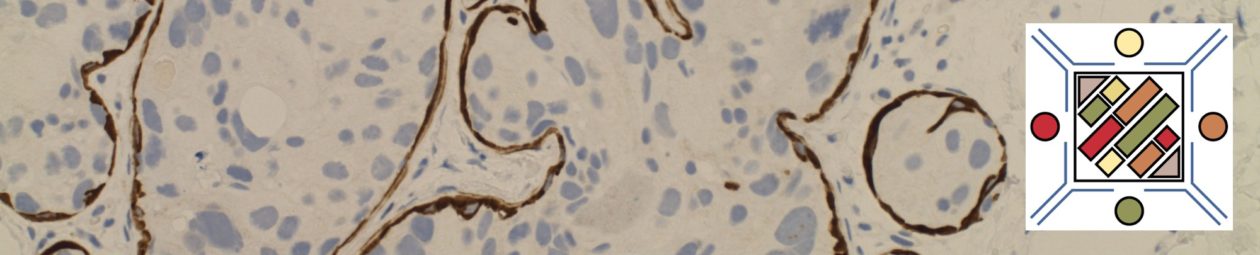
Unlike ALK, there is no known normal tissue counterpart which can be used as a control. Therefore, known ROS-1 positive tumors or cell lines (HCC78 cell line with the SLC34A2-ROS1 rearrangement) are generally used. ROS-1 expression is cytoplasmic with described expression ranging from finely granular to globular cytoplasmic staining and membranous staining. No consensuses has been established as to the minimal threshold of positivity.
Possible interpretation pitfalls include weak staining of type II pneumocytes and alveolar macrophages along with osteoclast-type giant cells in bone biopsies. Like any immunostain, contextual evaluation is critical.
References
Thunnissen E, Allen TC, Adam J, Aisner DL, Beasley MB, Borczuk AC, et al. Immunohistochemistry of Pulmonary Biomarkers: A Perspective From Members of the Pulmonary Pathology Society. Arch Pathol Lab Med. 2018;142: 408–419. doi:10.5858/arpa.2017-0106-SA
Shaw AT, Ou S-HI, Bang Y-J, Camidge DR, Solomon BJ, Salgia R, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 2014;371: 1963–1971. doi:10.1056/NEJMoa1406766
Bubendorf L, Büttner R, Al-Dayel F, Dietel M, Elmberger G, Kerr K, et al. Testing for ROS1 in non-small cell lung cancer: a review with recommendations. Virchows Arch. 2016;469: 489–503. doi:10.1007/s00428-016-2000-3
Boyle TA, Masago K, Ellison KE, Yatabe Y, Hirsch FR (2015) ROS1 immunohistochemistry among major genotypes of non- small-cell lung cancer. Clin Lung Cancer 16(2):106–111. doi:10.1016/j.cllc.2014.10.003
CaoB,WeiP,LiuZ,BiR,LuY,ZhangL,ZhangJ,YangY,Shen C, Du X, Zhou X (2016) Detection of lung adenocarcinoma with ROS1 rearrangement by IHC, FISH, and RT-PCR and analysis of its clinicopathologic features. Onco Targets Ther 9:131–138. doi:10.2147/OTT.S94997
Sholl LM, Sun H, Butaney M, Zhang C, Lee C, Janne PA, Rodig SJ (2013) ROS1 immunohistochemistry for detection of ROS1-rearranged lung adenocarcinomas. Am J Surg Pathol 37(9):1441–1449. doi:10.1097/PAS.0b013e3182960fa7
Yoshida A, Tsuta K, Wakai S, Arai Y, Asamura H, Shibata T, Furuta, K, Kohno T, Kushima R (2014) Immunohistochemical detection of ROS1 is useful for identifying ROS1 rearrangements in lung can- cers. Mod Pathol 27(5):711–720. doi:10.1038/modpathol.2013.192
Rogers TM, Russell PA, Wright G, Wainer Z, Pang JM, Henricksen LA, Singh S, Stanislaw S, Grille J, Roberts E, Solomon B, Fox SB (2015) Comparison of methods in the detection of ALK and ROS1 rearrangements in lung cancer. J Thorac Oncol 10(4):611–618. doi:10.1097/JTO.0000000000000465
Rimkunas VM, Crosby KE, Li D, Hu Y, Kelly ME, Gu TL, Mack JS, Silver MR, Zhou X, Haack H (2012) Analysis of receptor tyro- sine kinase ROS1-positive tumors in non-small cell lung cancer: identification of a FIG-ROS1 fusion. Clin Cancer Res 18(16): 4449–4457. doi:10.1158/1078-0432.CCR-11-3351
Mescam-Mancini L, Lantuejoul S, Moro-Sibilot D, Rouquette I, Souquet PJ, Audigier-Valette C, Sabourin JC, Decroisette C, Sakhri L, Brambilla E, McLeer-Florin A (2014) On the relevance of a testing algorithm for the detection of ROS1-rearranged lung adenocarcinomas. Lung Cancer 83(2):168–173. doi:10.1016/j. lungcan.2013.11.019
Shan L, Lian F, Guo L, Qiu T, Ling Y, Ying J, Lin D (2015) Detection of ROS1 gene rearrangement in lung adenocarcinoma: comparison of IHC, FISH and real-time RT-PCR. PLoS One 10(3): e0120422. doi:10.1371/journal.pone.0120422